Automated Cell Cluster Detection
Mammary epithelial cells stained with Hoechst (blue) and an antibody recognizing HMGB1 (green). 9 x 9 stitched image of 20X images captured on a Nikon Ti-2 eclipse fluorescence microscope by Ilechukwu Agu
During paracrine signaling, cells send and receive ligands that trigger gene expression responses. As a result, cells express proteins in groups or “clusters” where nearby cells express a certain protein, while others do not.
Quantification of these clusters traditionally require manual identification and drawing of the outlines for each cluster. This is a time intensive method and requires painstaking effort by the researcher.
This script automatically counts the number of clusters in an image and quantifies characteristics of each cluster using image processing techqniques described below. This includes the number of cells in each cluster, area and location of each cluster. Furthermore, it quantifies the intensity of protein expression in each cluster.
This method drastically speeds up the analysis time and standardizes the workflow. This leads to less human error and more consistency acroess researchers and datasets.
This script can be used on any immunofluorescence of any protein!
Workflow
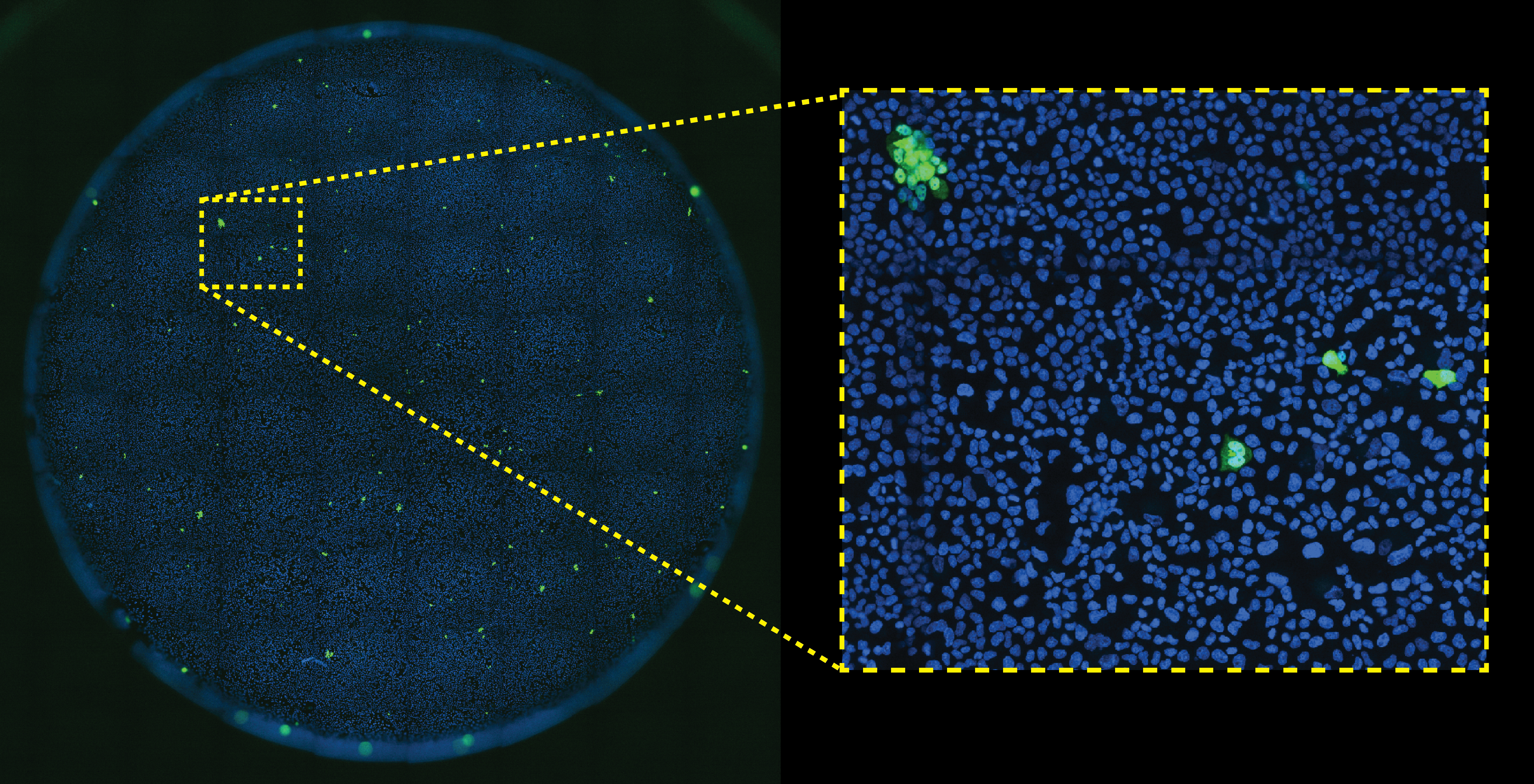
We will be looking at the whole image and a zoomed in region of the image.
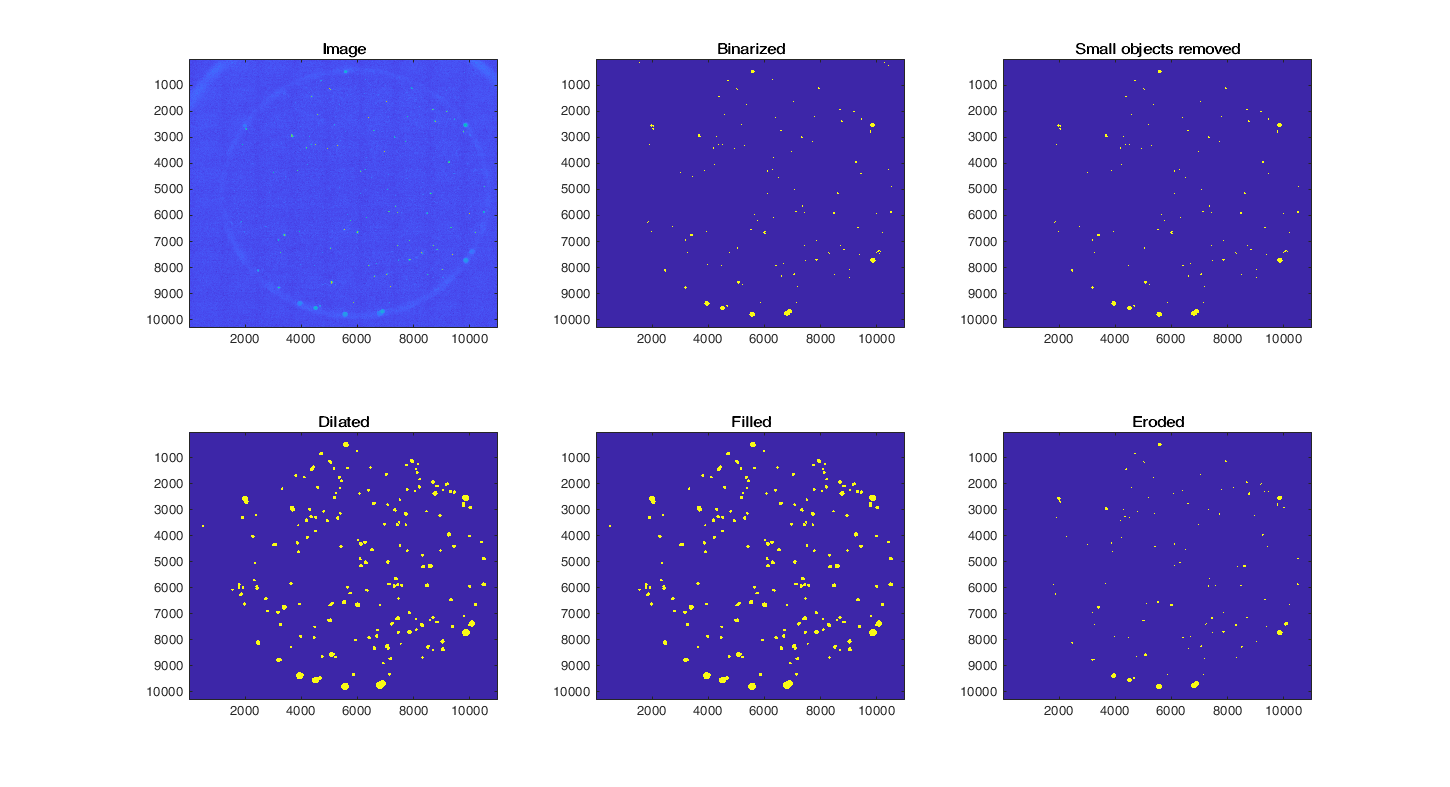
Object masks are generated from immunofluorescence images depicting HMGB1 (in green) and hoechst staining (in blue). The hoechst image undergoes nuclear segmentation to quantify the number of nuclei within each mask. To enhance detectability, images are binarized using a user-defined threshold (imbinarize), followed by the removal of small, noisy pixels (bwareaopen). The masks are then subjected to dilation and filling operations (imdilate and imfill) to achieve a smoother appearance without any holes. Then, the masks undergo an erosion step (imerode) to reverse the previous dilation process.
To count the number of cells in a cluster, object masks were overlaid with the nuclear segmentation resulting from Hoechst-based segmentation. Incorrect masks were filtered out by thresholding the min/max mask area and removing masks that did not contain any nuclei. Finally, each mask are counted and characterized to obtain pixel intensities (protein expression) and object areas.
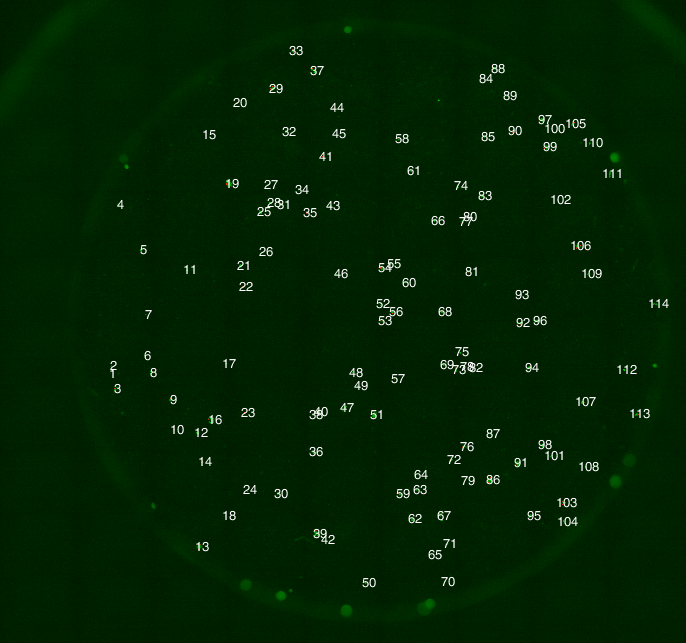
YFP channel image: counts overlayed onto each cluster
If we zoom into the image, we can visualize the mask created by the object segmentation.
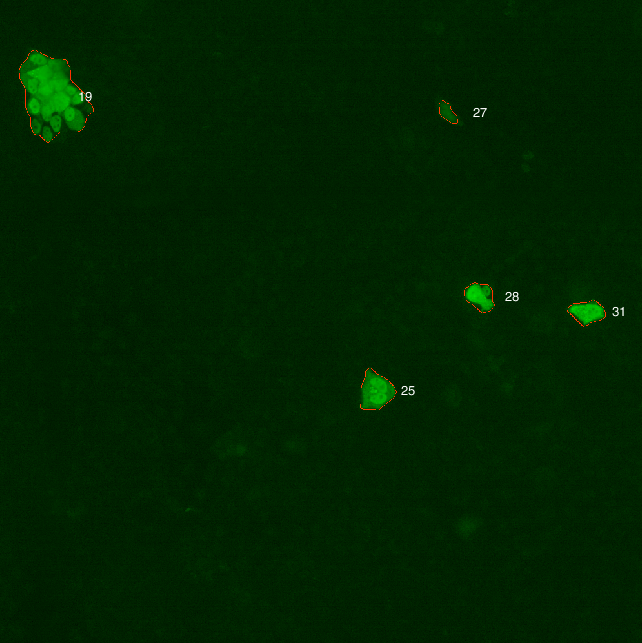
YFP channel image: Cluster (green) and mask overlay (red)
Nuclear segmentation
To count the number of cells in each plaque, nuclear segmentation is conducted in the blue channel image (hoechst stain).
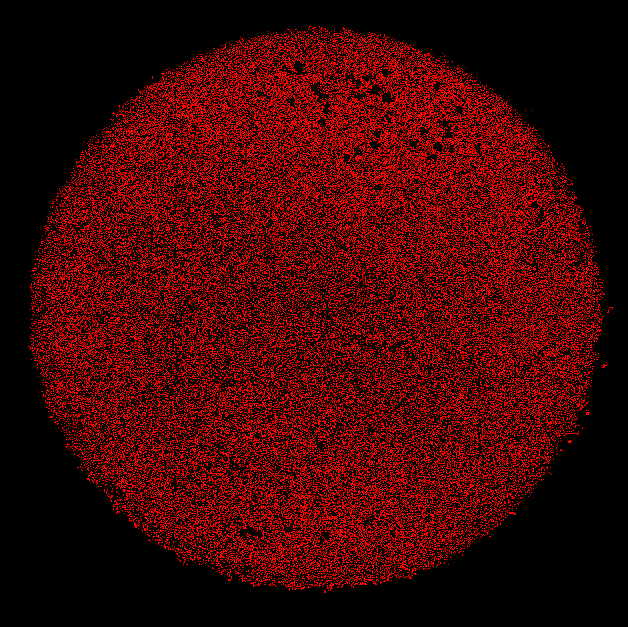
All segmented nuclear maks (red) |
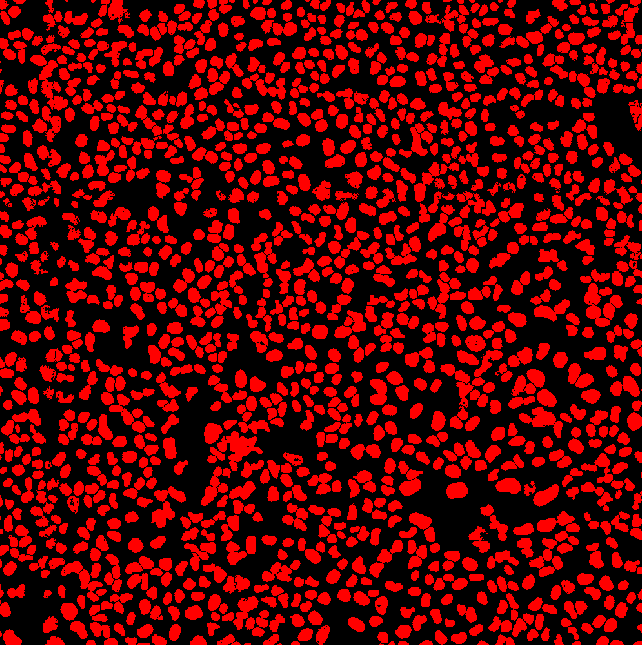
Zoomed in to our region of interest |
Then the cluster masks are overlayed onto the segmented nuclear masks, and the number of cells in mask is counted.
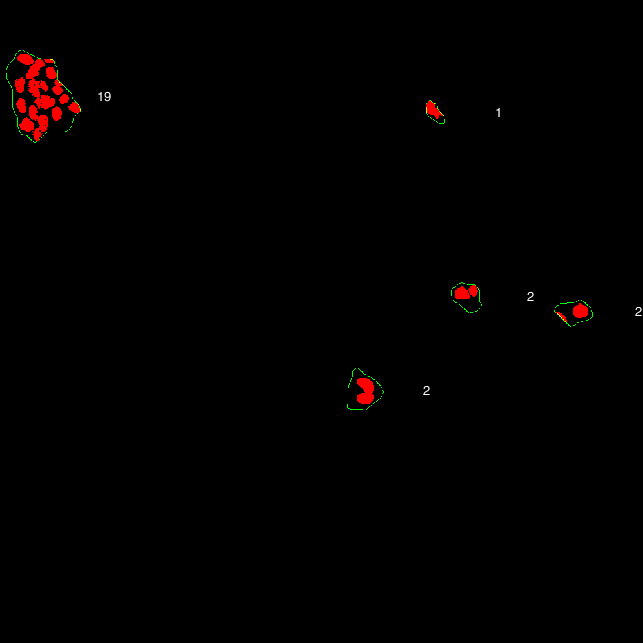
Nuclear masks (red) overlayed onto cluster masks (green). Number of cells within each mask are indicated.
This script allows us to count and properly quantify the clusters in each image! It will output the number of clusters, location, area and intensity of each cluster, and the number of cells in each cluster in a table format.