Spectral Unmixing
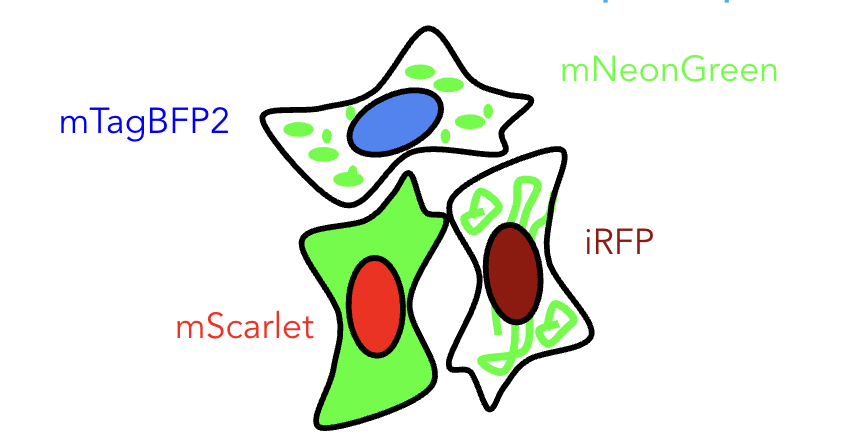
How do we image four different fluorescent proteins at once?
During my internship in the Leonetti group at Chan Zuckerberg Biohub, my project involved correcting for crosstalk in confocal microscopy images. My mentor and I configured the microscope for dual-camera imaging. This required the simultaneous excitation of two fluorescent proteins, and then using a beam-splitter to send low and high wavelength emission to different cameras, respectively. The configuration allowed for imaging of four different fluorescent proteins in a sample.
The main challenge with this system was that some emitted light from one fluorescent protein may bleed through to the undesired camera, thereby resulting in crosstalk.
To overcome this, our approach was to model and simulate the light path of the microscope and calculate fluorophore sensitivity constants for a given set of fluorescent proteins in each of the imaging channels.
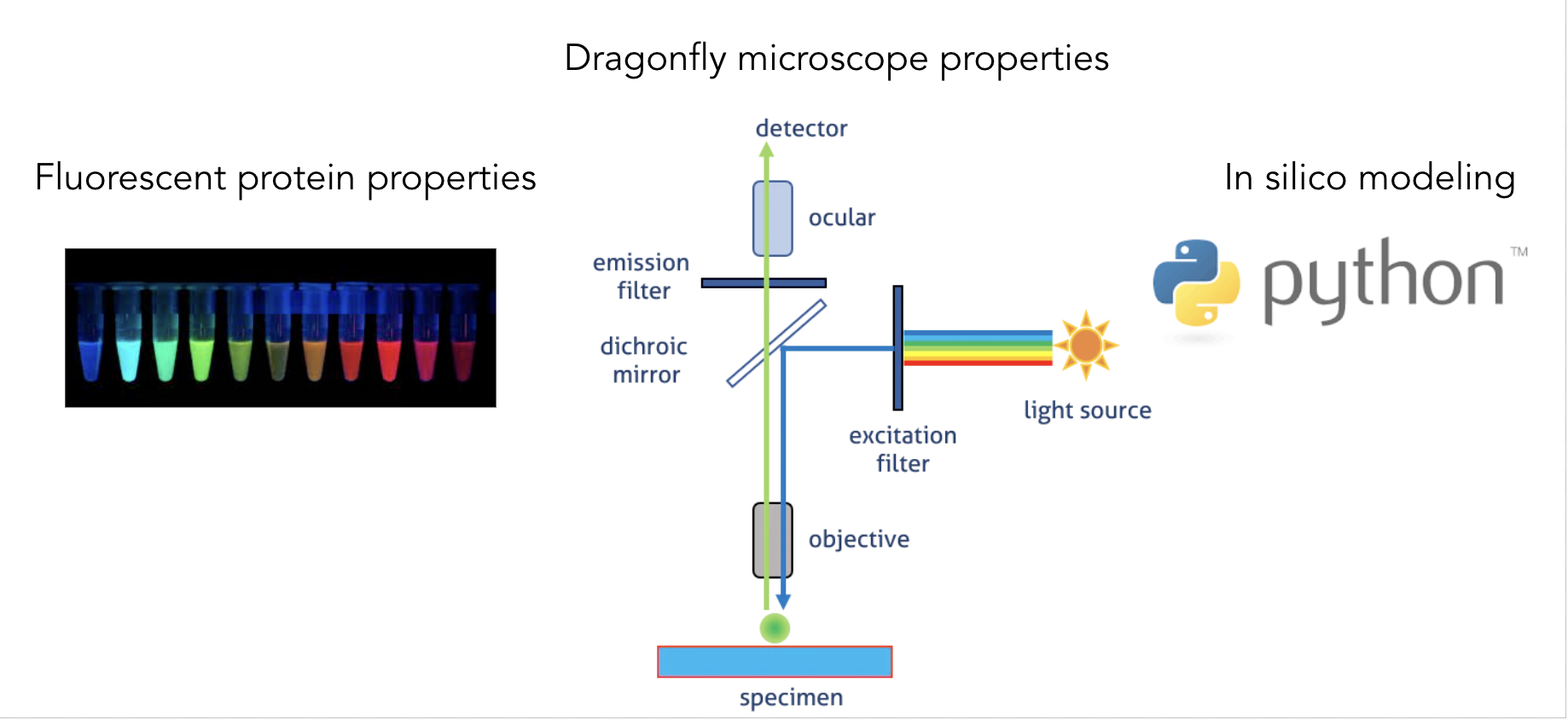
I used Python to create a function that inputs a set of fluorescent proteins and microscope acquisition settings to return the channel-fluorophore sensitivity constants, which then can be used to perform linear unmixing of pixel intensities of the captured image.
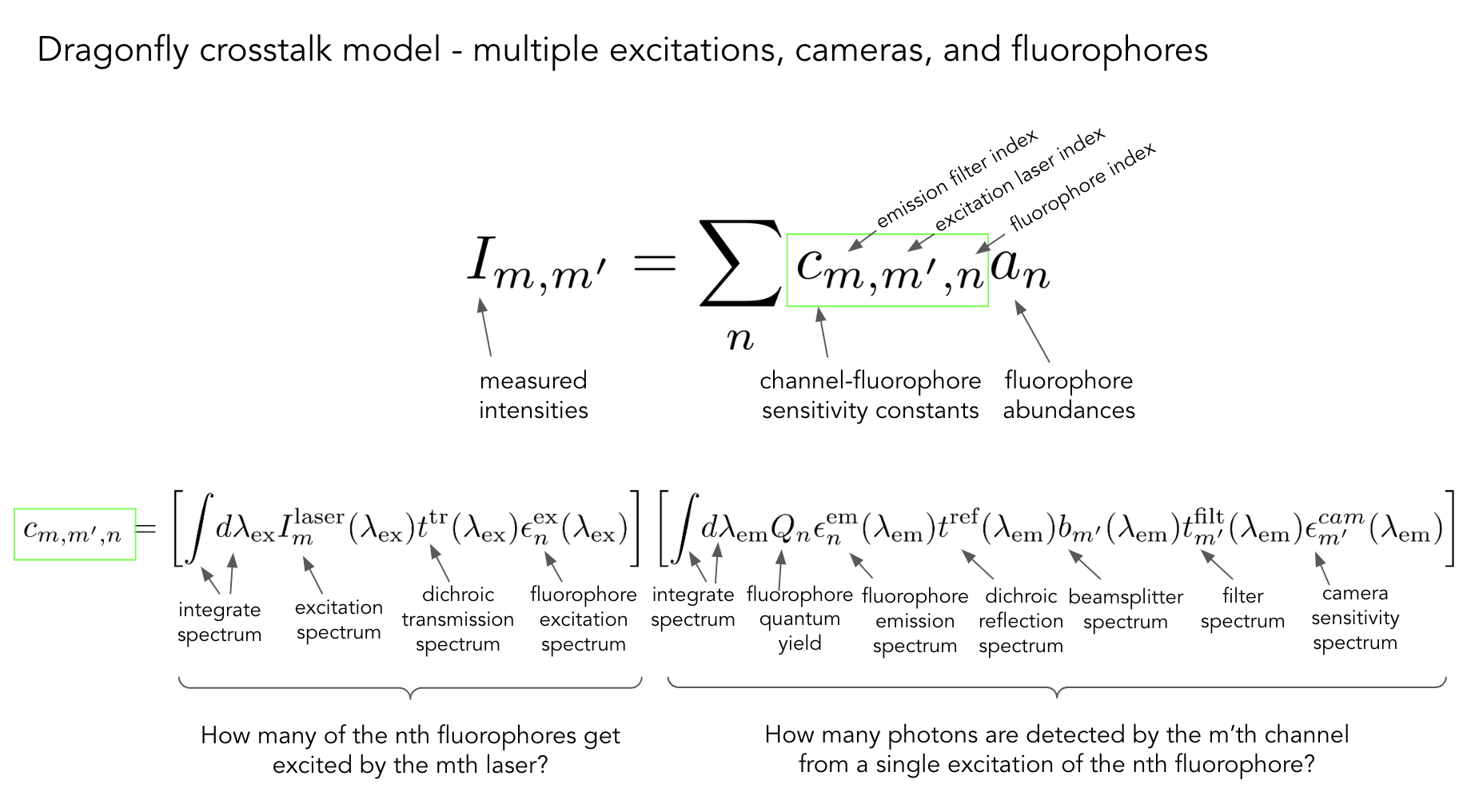
Our light path model
Imagine the following experimental setup: BFP and mScarlet imaged simultaneously on the left camera, along with mNeonGreen and iRFP760 simultaneously on the right camera. The model predicted that 1% of the signal in the red channel would be emission from BFP, and 16% of the signal in the far red will be emitted from mScarlet.
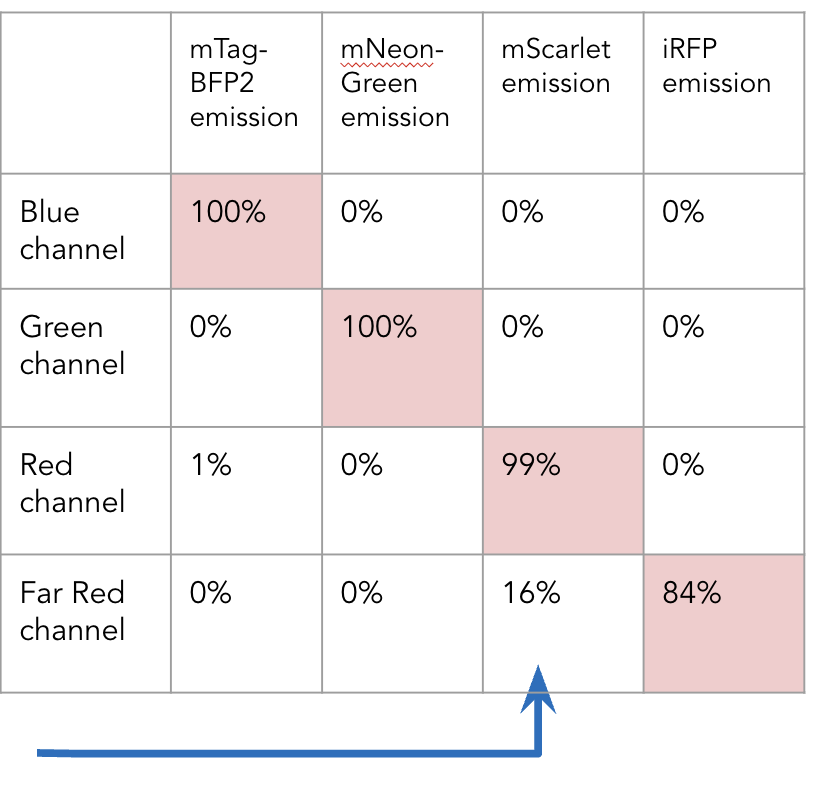
Light path simulation of BFP and mScarlet | mNeonGreen and iRFP760
We could then linearly unmix an image to correct for the crosstalk.
How it works
The spectral model calculates the expected signal intensities using the specfic configuration of the microscope, and the properties of each fluorescent proteins requested. The shaed region below is the product of all of the listed properties, summed across wavelengths.
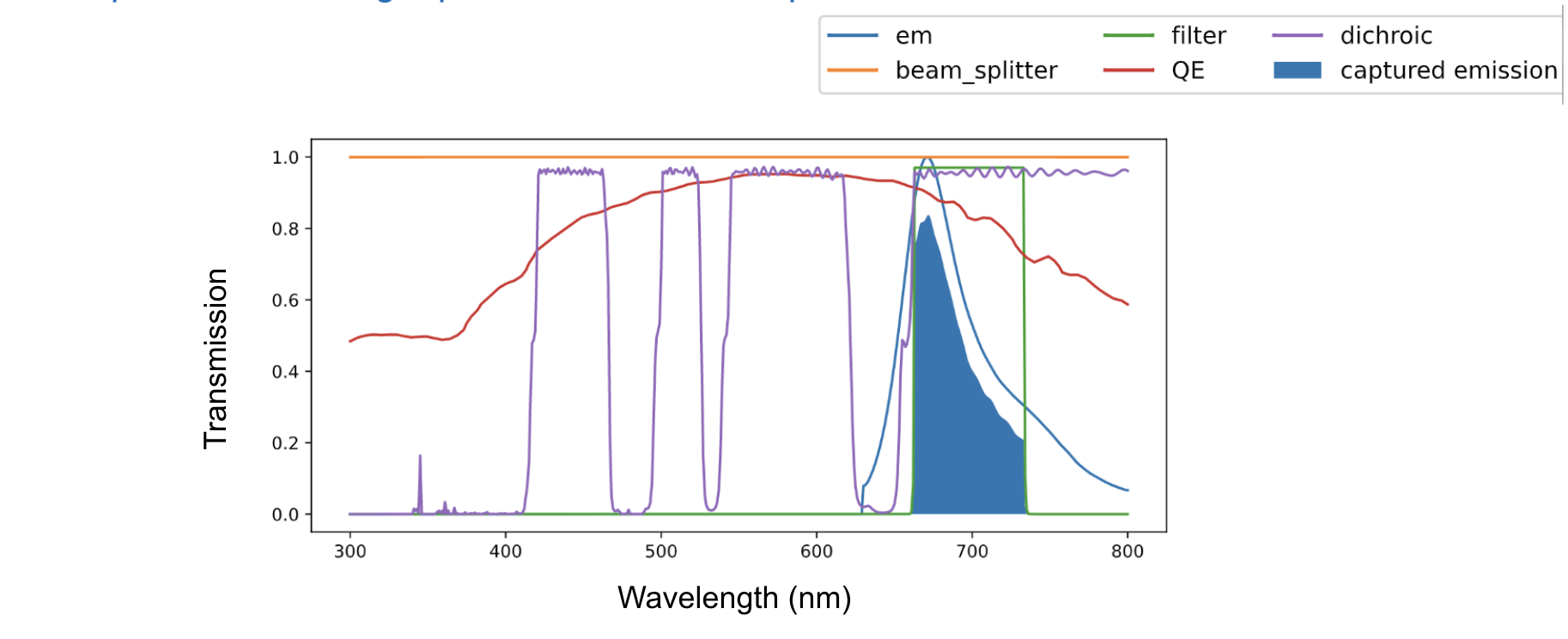
Expected signal calculation for iRFP670
The python code will do this calculatation for each input fluorophore in each channel. Here’s an example input and the plotted spectra:
# Define fluorescent proteins
FPs = [['mTagBFP2','mScarlet'],['mNeonGreen','iRFP670']]
# Define miscroscope laser settings
exc_lines = [['405','561'],['488','637']]
laser_powers = [1,1,1,1]
exposure_times = [100, 200]
#calculate unmixing matrix
c_2d = util.specmix_matrix(datafolder,FPs, exc_lines, laser_powers, exposure_times)
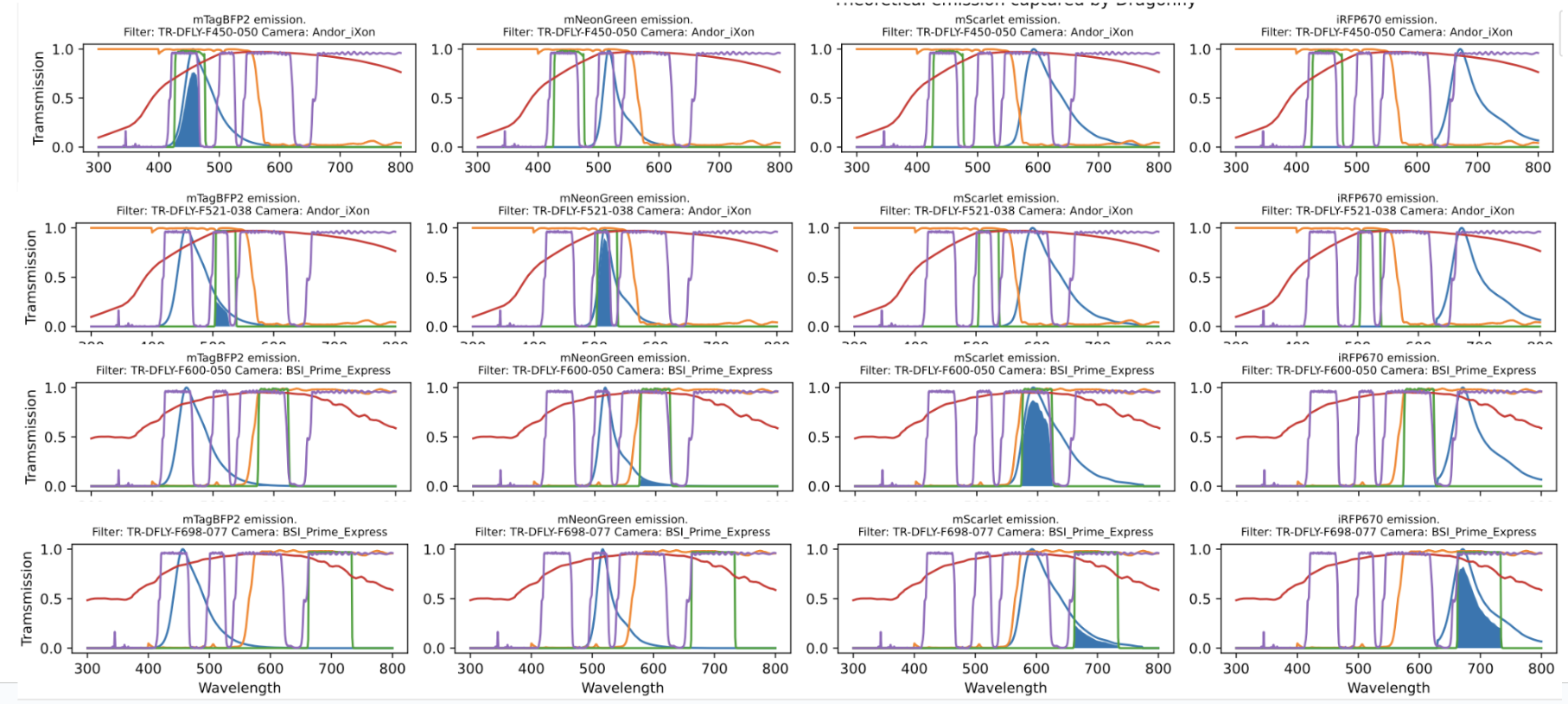
Four requested fluorophores in four channels
The script will output these shaded values as a normalized matrix which gives the user the expected signal per fluorophore per channel as percentages. These values can then be used to correct for crosstalk in the images taken on this particular microscope given these specific acquisition settings.